Publications研究業績
Papers Reviews Talks Patents
Preprint
79. *Harashima T, *Iino R
Artificial DNA-nano/microparticle motors: Factors governing speed, run-length, and unidirectionality revealed by geometry-based kinetic simulations
bioRxiv DOI:10.64898/2026.02.11.705463
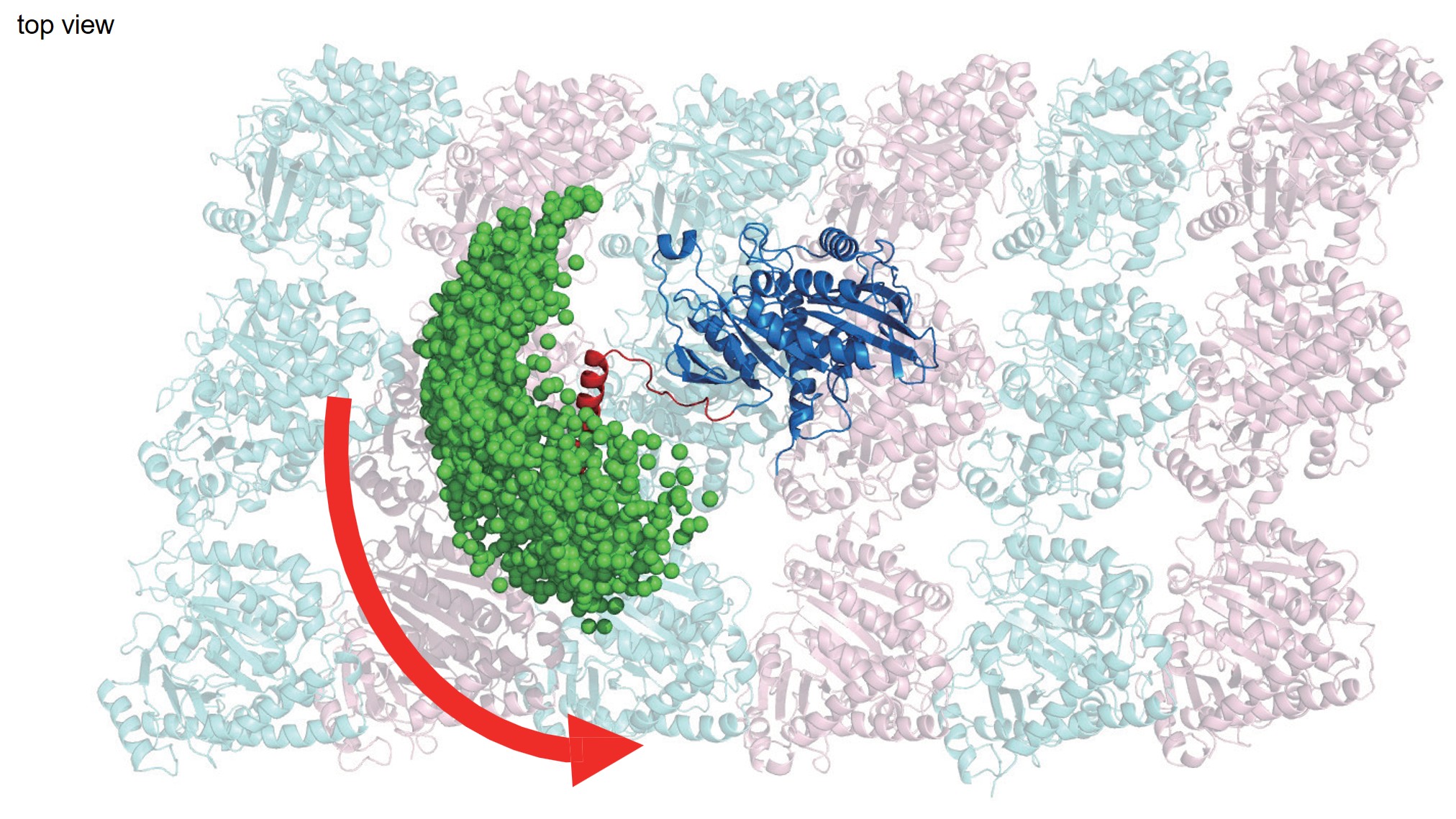
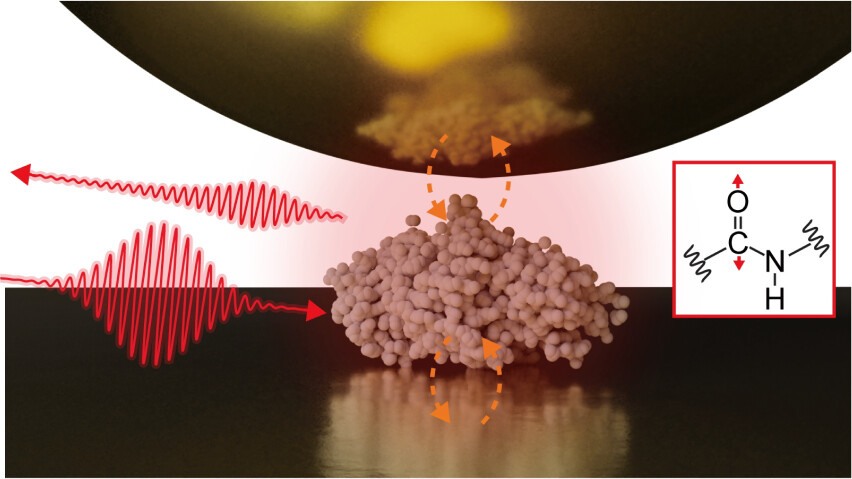
78. Fukuda S, Otomo A, Iino R, *Ando T
Enhancing the imaging rate of high-speed atomic force microscopy using a combination of multiple techniques
bioRxiv DOI:10.64898/2025.12.22.695957
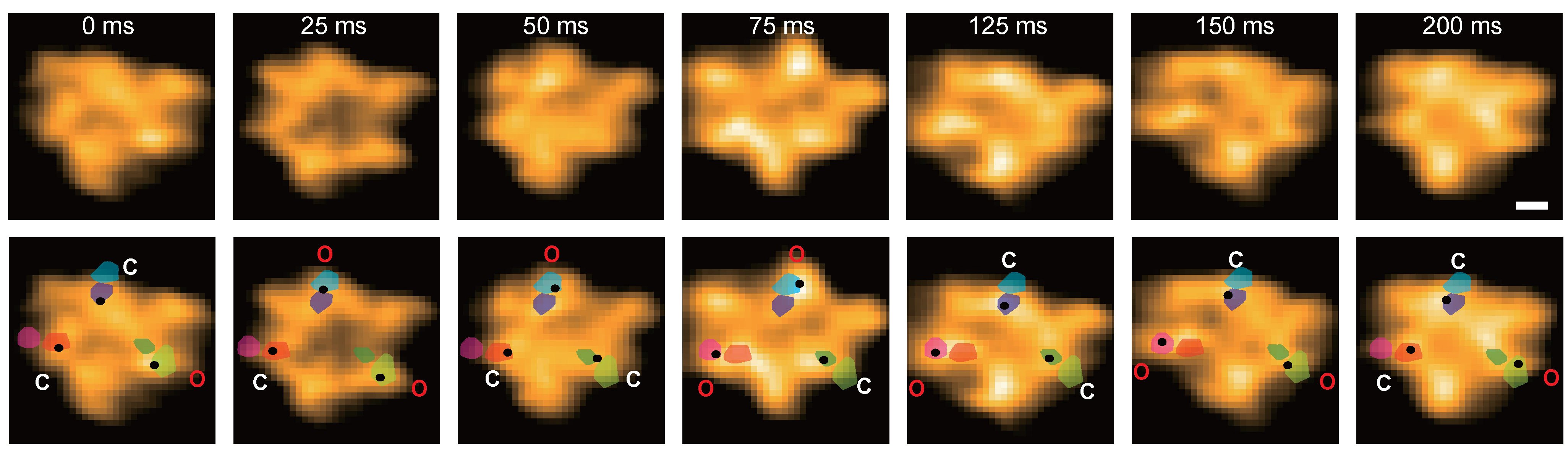
77. Tanimoto H, Tsudome M, Tachioka M, Baba A, Miyazaki M, Uchihashi T, Iino R, *Deguchi S, *Nakamura A
A dual-domain chitinase mechanism enables marine bacteria to sense and localize sparse crystalline chitin in the ocean
bioRxiv DOI:10.64898/2025.12.08.690172
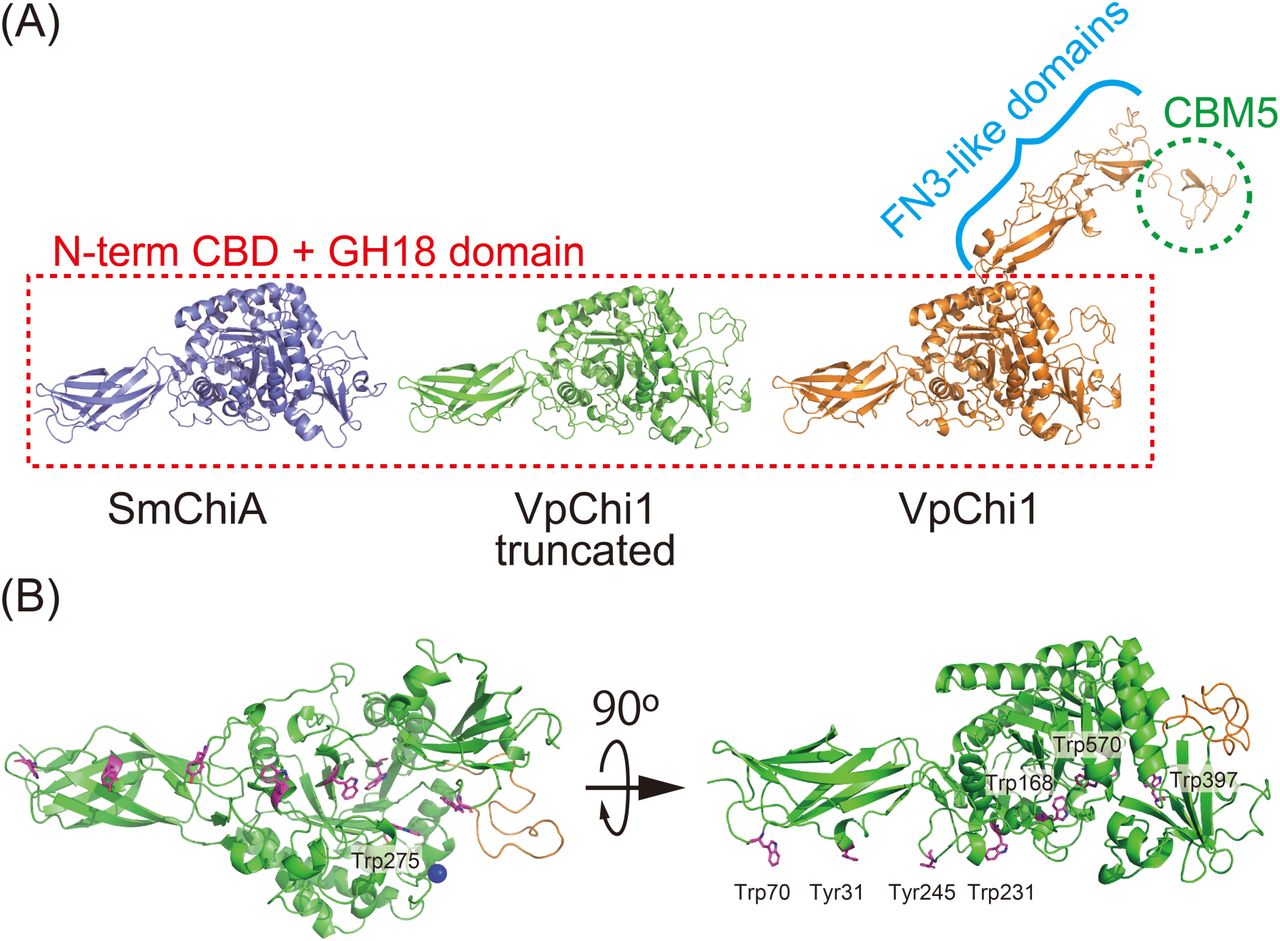
76. *Chong SH, *Iino R
Neck-region–microtubule interactions direct counterclockwise stepping of kinesin-1
bioRxiv DOI:10.1101/2025.06.07.658466
2025
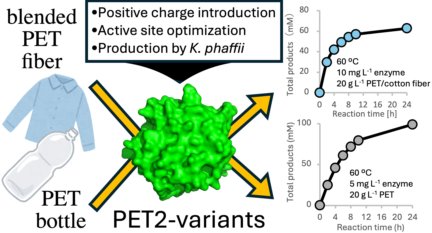
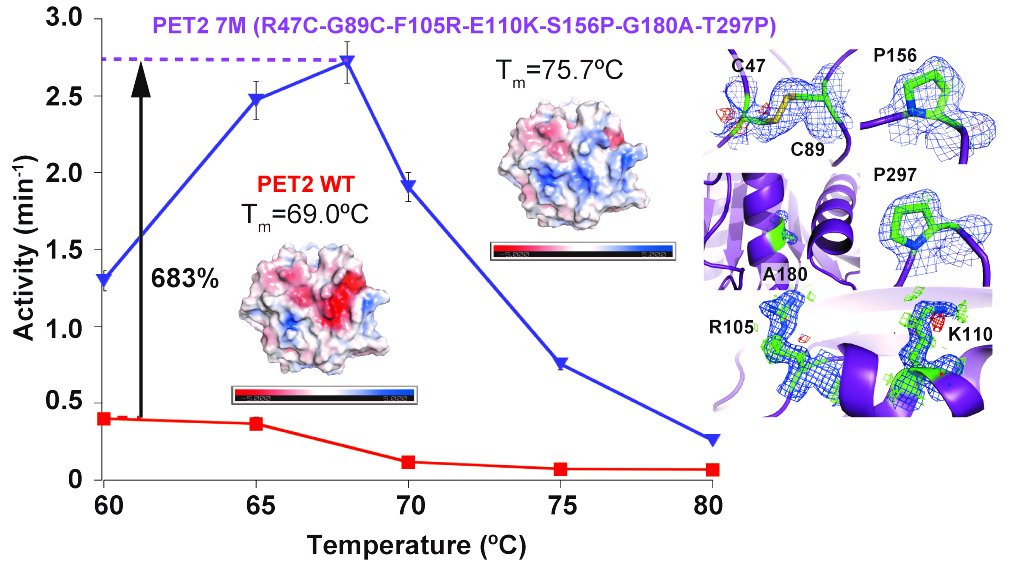
75. #Matsuzaki T, #Saeki T, Yamazaki F, Koyama N, Okubo T, Hombe D, Ogura Y, Hashino Y, Tatsumi-Koga R, Koga N, Iino R, *Nakamura A (#Equal contribution)
Development and production of moderate-thermophilic PET hydrolase for PET bottle and fiber recycling
ACS Sustainable Chem Eng 13, 10404–10417 (2025) DOI:10.1021/acssuschemeng.5c01602
Press release: Breakthrough engineered enzyme for recycling of PET bottle and blended fibers at moderate temperatures
研究成果: 改良型酵素で50~60 ℃の比較的低い温度でのPETボトルや混紡繊維の分解に成功
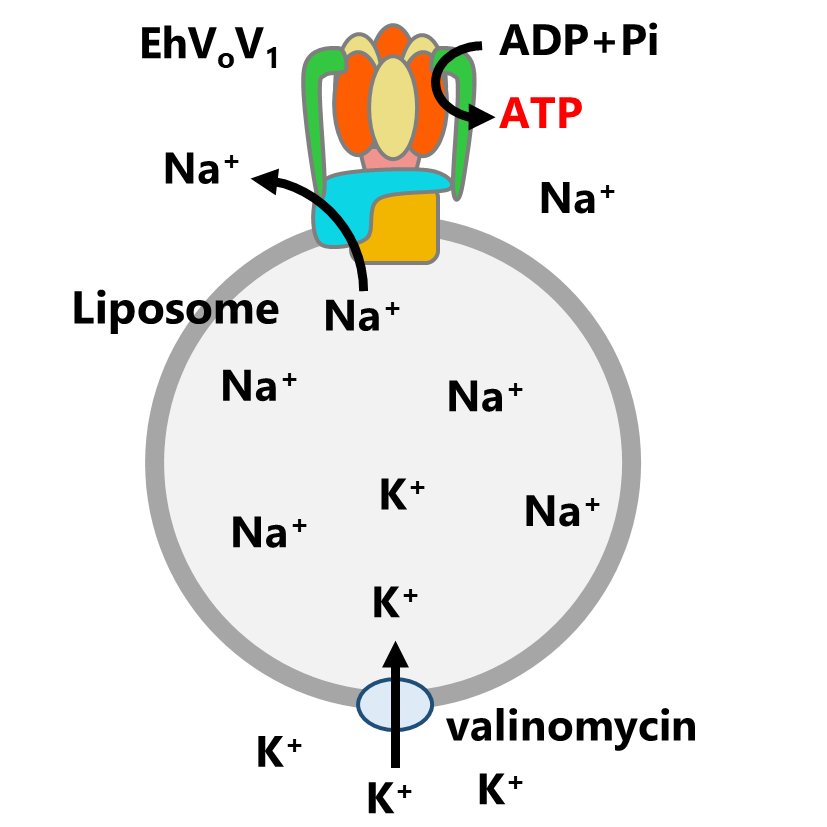
74. *Otomo A, Zhu GHL, Okuni Y, Yamamoto M, *Iino R
ATP synthesis of Enterococcus hirae V-ATPase driven by sodium motive force
J Biol Chem 301, 108422 (2025) DOI:10.1016/j.jbc.2025.108422
bioRxiv DOI:10.1101/2024.12.26.630461
概要解説音声
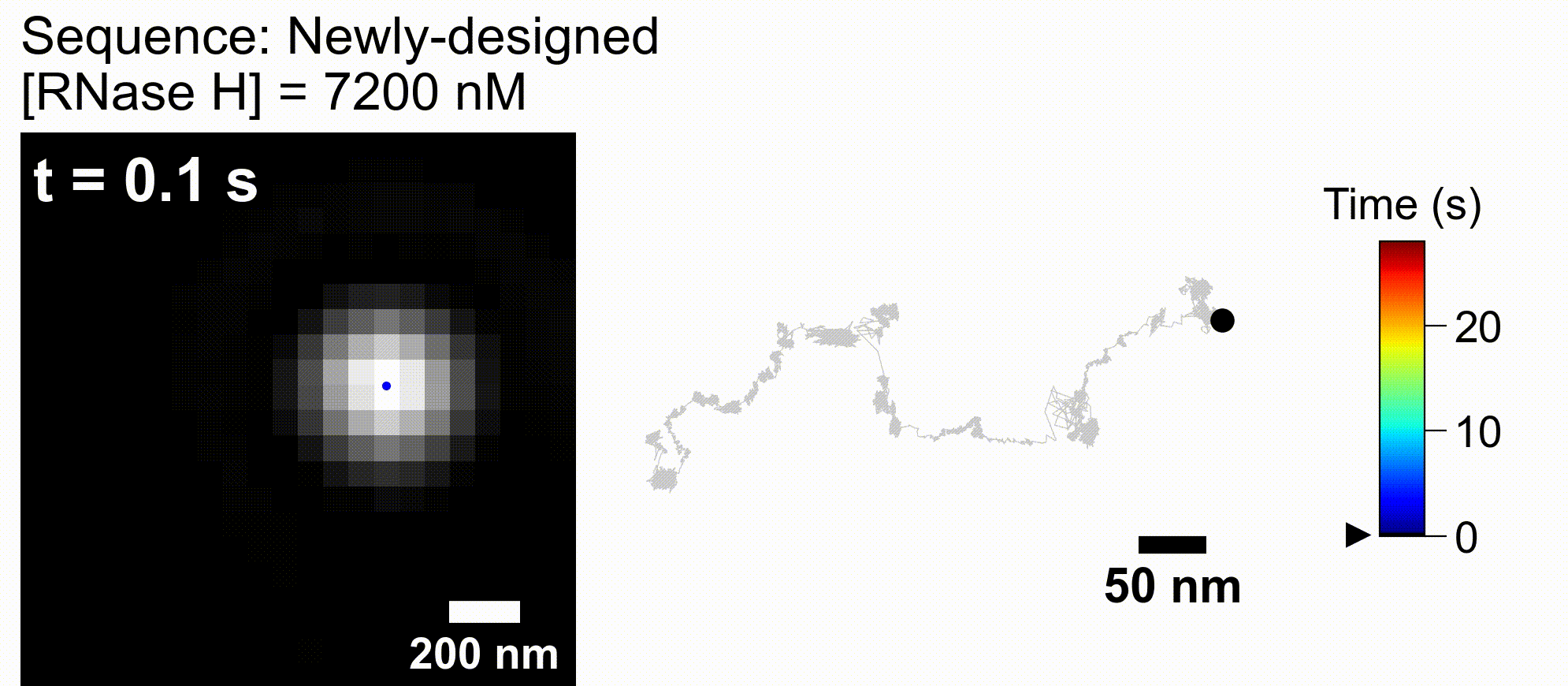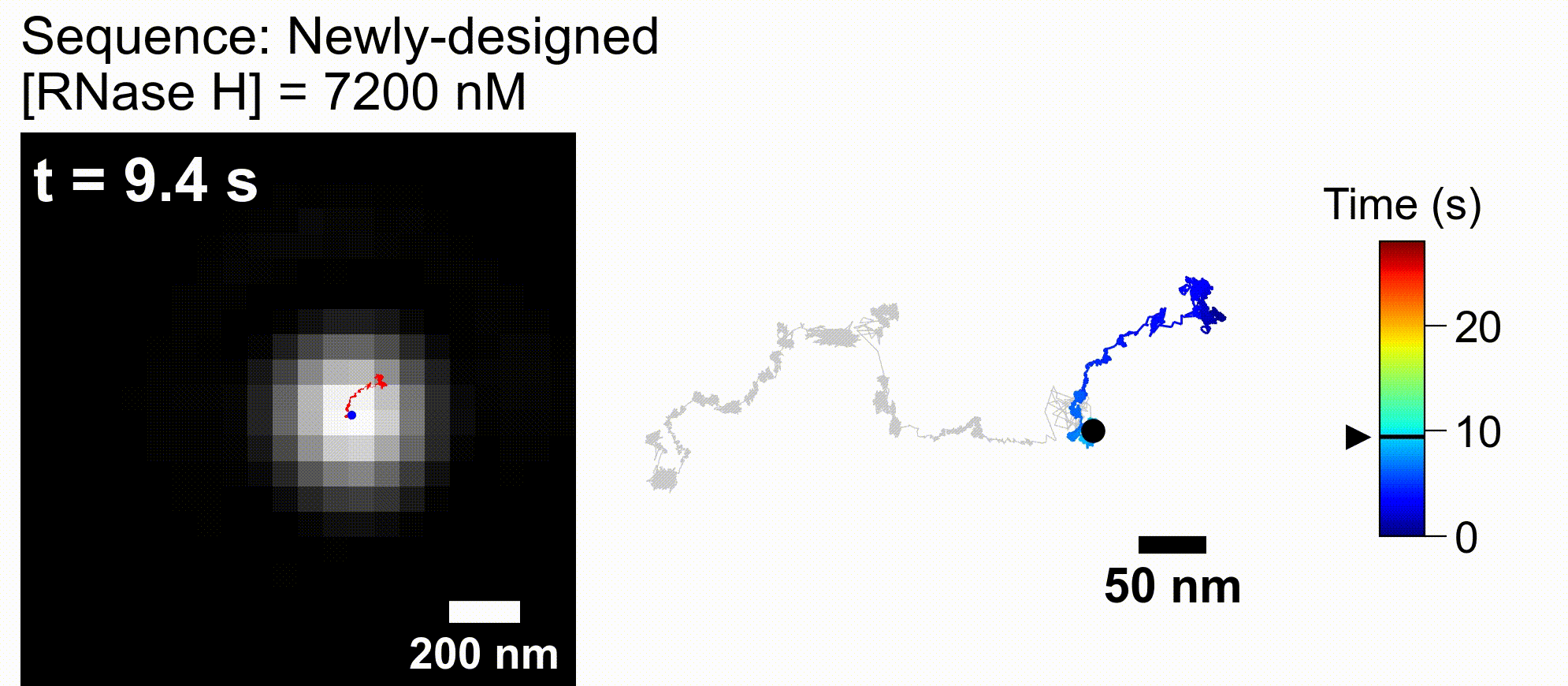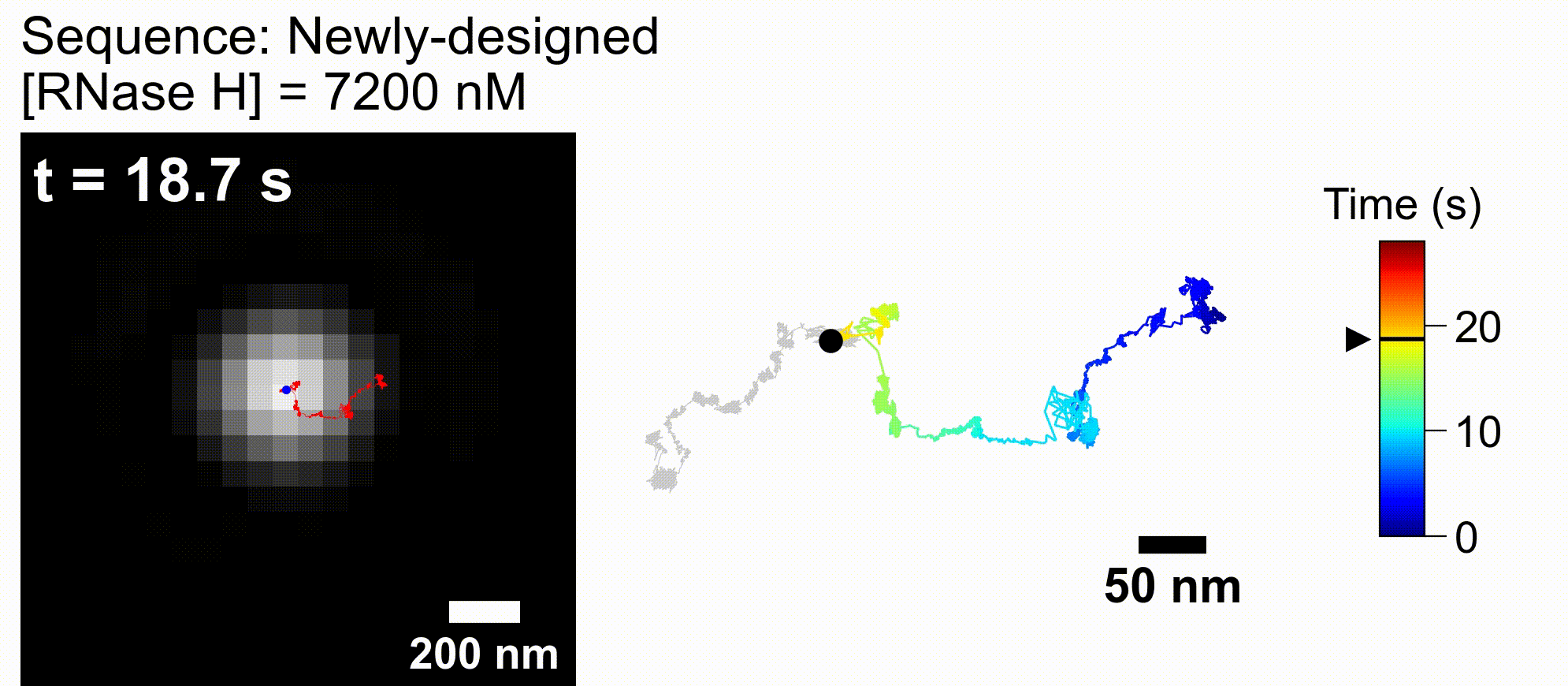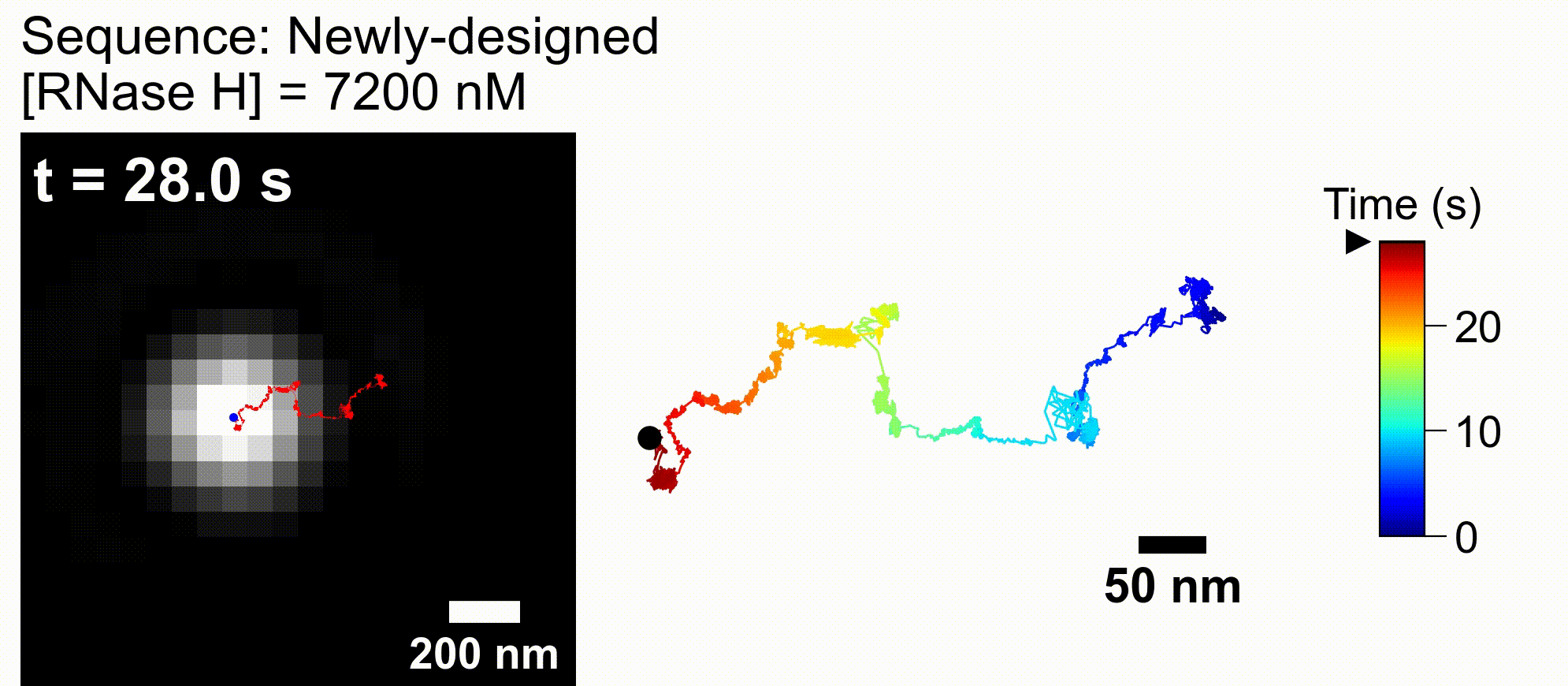
73. *Harashima T, Otomo A, *Iino R
Rational engineering of DNA-nanoparticle motor with high speed and processivity comparable to motor proteins
Nat Commun 16, Article number 729 (2025) DOI:10.1038/s41467-025-56036-0
bioRxiv DOI:10.1101/2024.05.23.595615
概要解説音声
Press release: Can DNA-nanoparticle motors get up to speed with motor proteins?
プレスリリース:人工分子モーターの合理的な改造で天然のモータータンパク質に匹敵する運動速度と走行距離を達成
ケムステ:モータータンパク質に匹敵する性能の人工分子モーターをつくる
科学新聞:天然のモータータンパク質に匹敵 運動速度と走行距離 人工分子モーターで実現(2025年2月21日付4面)
客观日本:日本分子研成功利用人工分子马达实现媲美天然马达蛋白的运动速度和距离,有望应用于高速运算和疾病检测领域


72. Suzuki K, Goto Y, Otomo A, Shimizu K, Abe S, Moriyama K, Yasuda S, Hashimoto Y, Kurushima J, Mikuriya S, Imai FL, Adachi N, Kawasaki M, Sato Y, Ogasawara S, Iwata S, Senda T, Ikeguchi M, Tomita H, Iino R, Moriya T, *Murata T
Na+-V-ATPase inhibitor curbs VRE growth and unveils Na+ pathway structure
Nat Struct Mol Biol 32, 450–458 (2025) DOI:10.1038/s41594-024-01419-y
プレスリリース: バンコマイシン耐性腸球菌(VRE)感染症の治療に道 -ナトリウムポンプ阻害剤の発見とその阻害機構を解明-
2024
71. Otomo A, Wiemann J, Bhattacharyya S, Yamamoto M, *Yu Y, *Iino R
Visualizing single V-ATPase rotation using Janus nanoparticles
Nano Lett 24, 15638–15644 (2024) DOI:10.1021/acs.nanolett.4c04109
bioRxiv DOI:10.1101/2024.08.22.609254
論文紹介:ヤヌスナノ粒子を用いたV-ATPaseの1分子計測(分子サイバネNL原稿)

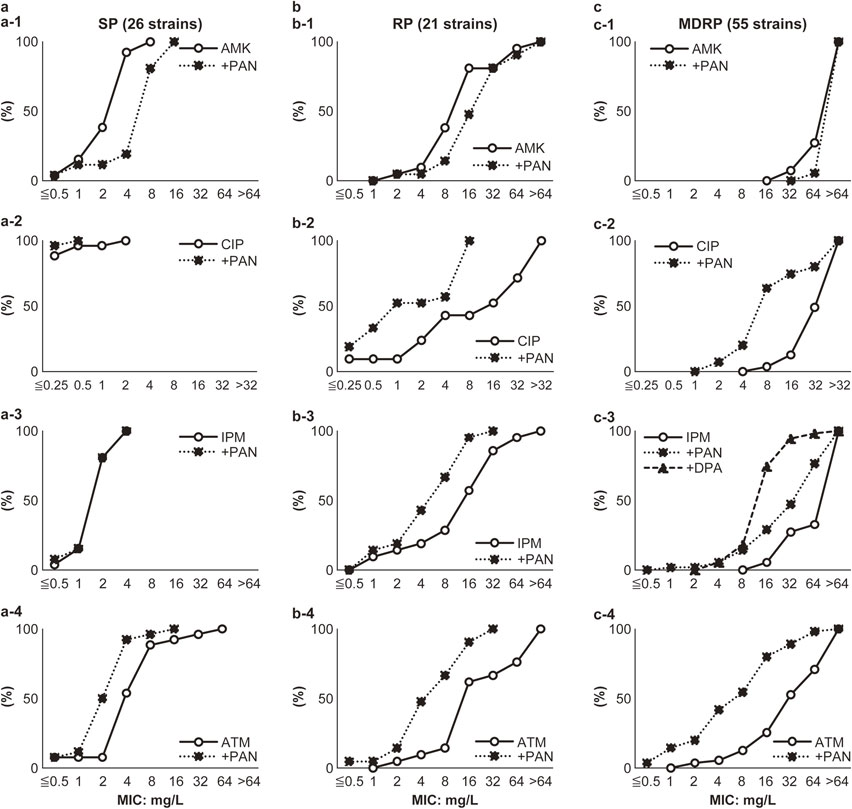
70. Matsumoto Y, Yamasaki S, Hayama K, Iino R, Noji H, Yamaguchi A, *Nishino K
Changes in the expression of mexB, mexY, and oprD in clinical Pseudomonas aeruginosa isolates
Proc Jpn Acad Ser B 100, 57-67 (2024) DOI:10.2183/pjab.100.006
69. *Nishida J, Otomo A, Koitaya T, Shiotari A, Minato T, Iino R, and *Kumagai T
Sub-tip-radius near-field interactions in nano-FTIR vibrational spectroscopy on single proteins
Nano Lett 24, 836–843 (2024) DOI:10.1021/acs.nanolett.3c03479
Press release: Observing single protein with infrared nanospectroscopy
プレスリリース>: 赤外光で単一のタンパク質を見る新技術
オプトロニクスオンライン:分子研,赤外光で単一のタンパク質を観察
2023
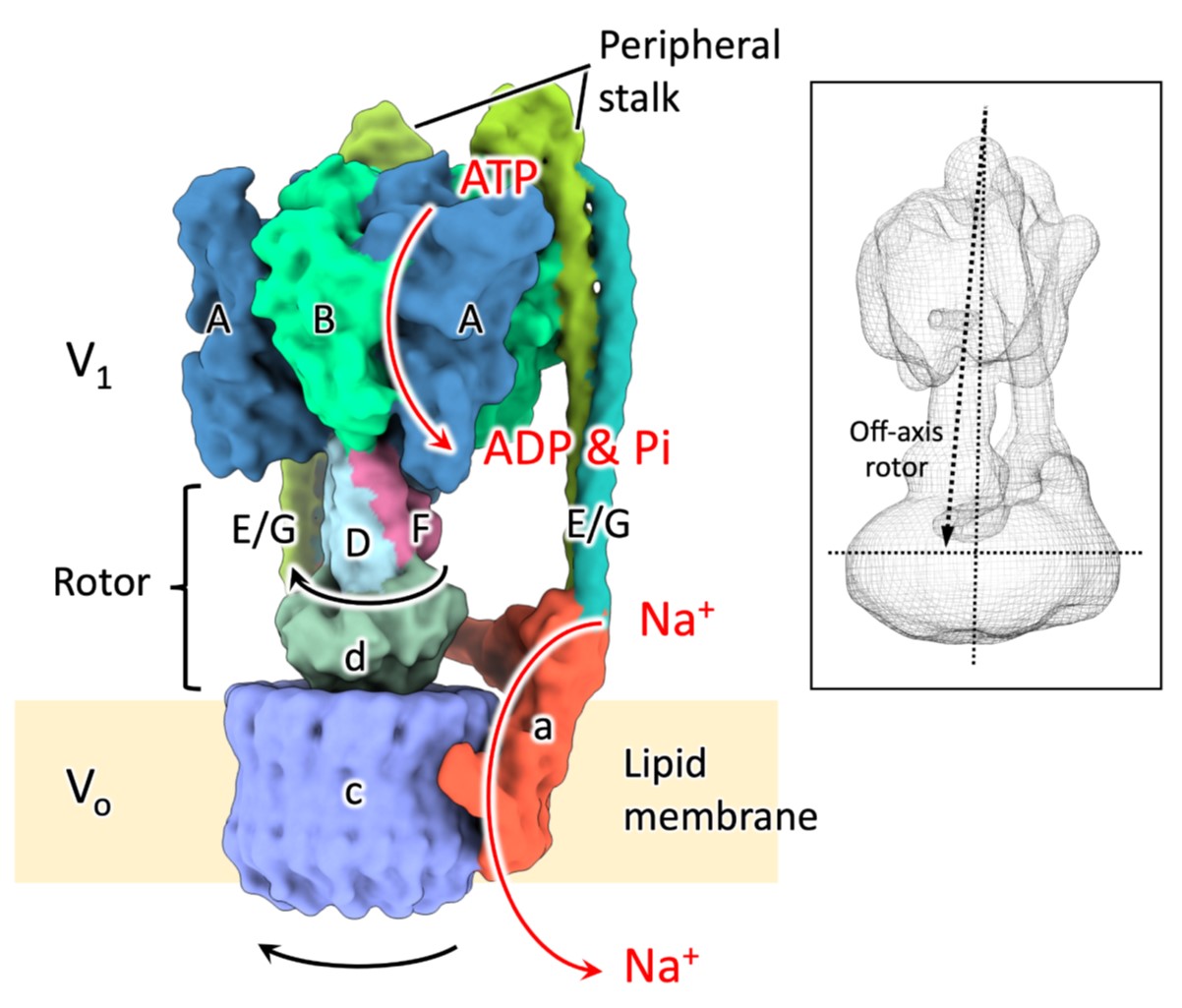
68. Burton-Smith RN, Song C, Ueno H, Murata T, Iino R, *Murata K
Six states of Enterococcus hirae V-type ATPase reveals non-uniform rotor rotation during turnover
Commun Biol 6, Article number 755 (2023) DOI:10.1038/s42003-023-05110-8
bioRxiv (2022) DOI:10.1101/2022.08.09.503272
Press release>: The structures of six states of a rotary sodium ion pump are revealed
プレスリリース:回転式ナトリウムイオンポンプの6つの中間構造すべてを立体構築することに成功
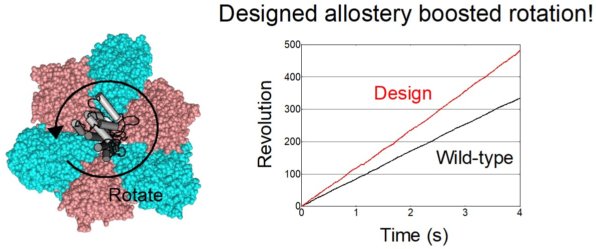
67. *Kosugi T, Iida T, Tanabe M, Iino R, *Koga N
Design of allosteric sites into rotary motor V1-ATPase by restoring lost function of pseudo-active sites
Nature Chemistry 15, 1591–1598 (2023) DOI:10.1038/s41557-023-01256-4
bioRxiv (2020) DOI: 10.1101/2020.09.09.288571
Press release: Scientists develop strategy to engineer artificial allosteric sites in protein complexes
プレスリリース:進化の過程で失った機能を復活させ、回転型分子モーターの加速に成功 ~タンパク質複合体の協奏的機能を制御する新手法~
日本経済新聞:分子科学研と生命創成探究センターなど、失った機能を計算機を用いたタンパク質設計で復活させ回転型分子モーターの加速に成功
2022
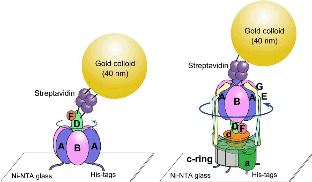
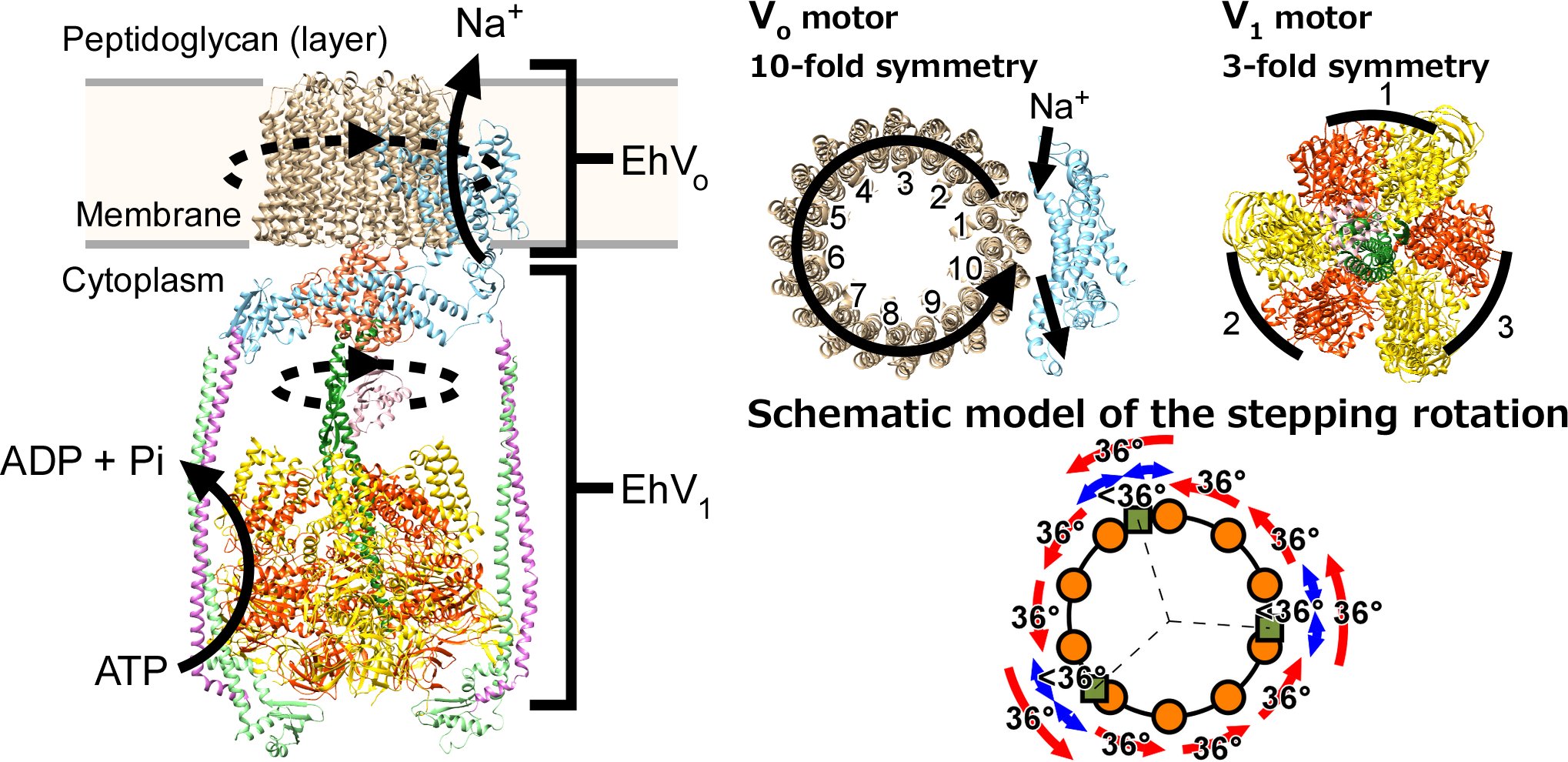
66. Otomo A, Iida T, Okuni Y, Ueno H, Murata T, *Iino R
Direct Observation of Stepping Rotation of V-ATPase Reveals Rigid Component in Coupling between Vo and V1 Motors
Proc Natl Acad Sci USA 119, e2210204119 (2022) DOI: 10.1073/pnas.2210204119
bioRxiv (2022) DOI: 10.1101/2022.06.13.494302
Press release:Molecular-motor specialists deepen our understanding of a rotary ion pump of the cell
プレスリリース:回転イオンポンプの2つの分子モーターは硬く繋がり連動して働く
2021
65. *Nakamura A, Kobayashi N, Koga N, *Iino R
Positive charge introduction on the surface of thermostabilized PET hydrolase facilitates PET binding and degradation
ACS Catalysis 11, 8550-8564 (2021) DOI:10.1021/acscatal.1c01204
Press release "Engineered protein inspired by nature may help plastic plague"
プレスリリース1: プラスチック分解酵素の熱安定性と活性を向上させ、熱安定性向上の構造的基盤と活性向上の機構を解明(2021年7月1日)
科学新聞:プラスチック分解酵素 熱安定性と活性を向上(2021年7月23日)
プレスリリース2: “酵素でペットボトルをリサイクル” PET分解酵素の産業応用に向けた共同研究を開始(2022年1月18日)
Press release 2: Joint Research Begins on PET-degrading Enzymes for Sustainable Recycling (2022/1/31)
日経バイオテク:キリンと静岡大、自然科学研究機構、酵素によるPETリサイクル技術を共同研究(2022年1月20日)
日刊工業新聞(PDF):酵素でペットボトル再生 キリン静岡大など共同研究(2022年1月20日24面、転載許諾済)
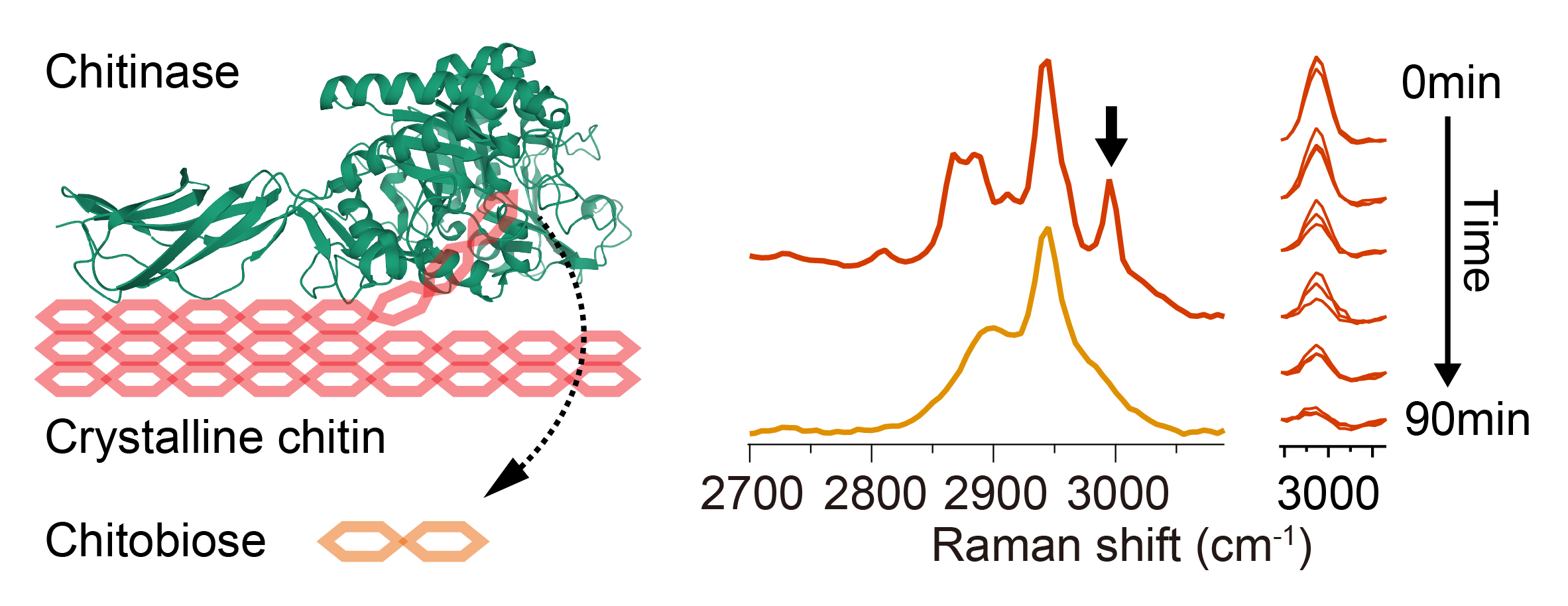
64. Ando J, Kawagoe H, Nakamura A, Iino R, *Fujita K
Label-free monitoring of crystalline chitin hydrolysis by chitinase based on Raman spectroscopy
Analyst 146, 4087-4094 (2021) DOI:10.1039/D1AN00581B
2020
63. Visootsat A, Nakamura A, Wang T-W, *Iino R
Combined approach to engineer a highly active mutant of processive chitinase hydrolyzing crystalline chitin
ACS Omega 5, 26807–26816 (2020) DOI:10.1021/acsomega.0c03911 OA
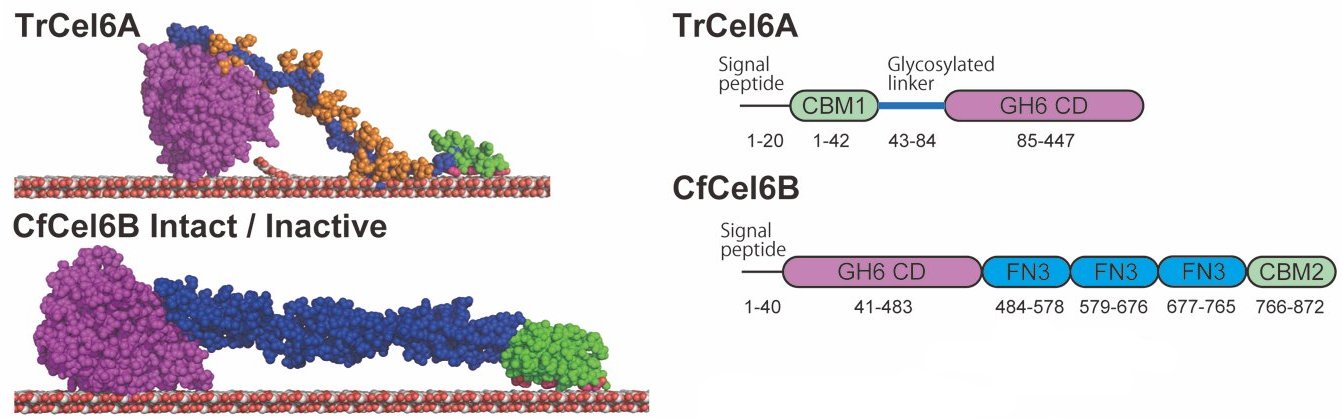
62. *Nakamura A, Ishiwata D, Visootsat A, Uchiyama T, Mizutani K, Kaneko S, Murata T, Igarashi K, *Iino R
Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases
J Biol Chem 295, 14606-14617 (2020) DOI:10.1074/jbc.RA120.014792 OA
Press release in EurekAlert
プレスリリース:目的は同じでも手段は異なる:細菌とカビのセロビオヒドロラーゼが結晶性セルロースを連続的に分解する戦略の違いを解明
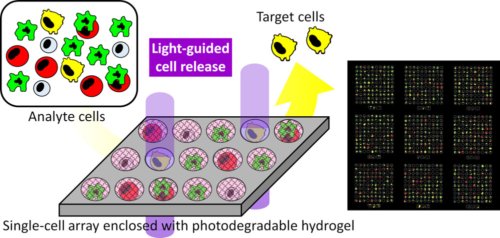
61. *Yamaguchi S, Takagi R, Hosogane T, Ohashi Y, Sakai Y, Sakakihara S, Iino R, Tabata KV, Noji H, *Okamoto A
Single cell array enclosed with a photodegradable hydrogel in microwells for image-based cell classification and selective photorelease of cells
ACS Appl. Bio Mater 3, 5887–5895 (2020) DOI:10.1021/acsabm.0c00583
60. *Okazaki K, Nakamura A, Iino R
Chemical-state-dependent free energy profile from single-molecule trajectories of biomolecular motor: Application to processive chitinase
J Phys Chem B 124, 6475–6487 (2020) DOI:10.1021/acs.jpcb.0c02698
Press release in EurekAlert
プレスリリース:分子モーターの1方向性運動モデルの新規推定法の開発 ―キチナーゼの運動メカニズム解明につながる―
科学新聞:分子モーターの1方向性運動:推定法の開発 分子研など成果(2020年8月28日6面)

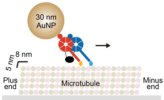
59. Ando J, Shima T, Kanazawa R, Shimo-Kon R, Nakamura A, Yamamoto M, Kon T, *Iino R
Small stepping motion of processive dynein revealed by load-free high-speed single-particle tracking
Sci Rep 10, Article number 1080 (2020) DOI:10.1038/s41598-020-58070-y OA
Press release in EurekAlert
プレスリリース:二本足のリニア分子モーターダイニンは小さな歩幅でふらふら歩く
オプトロニクスオンライン:分子研ら,レーザー顕微鏡で分子モーターを1分子観察 (2020年1月27日)
科学新聞:リニア分子モーター「ダイニン」 二足で歩行「まるで酔っ払い」(2020年2月14日6面)
58. Visootsat A, Nakamura A, Vignon P, Watanabe H, Uchihashi T, *Iino R
Single-molecule imaging analysis reveals the mechanism of a high-catalytic-activity mutant of chitinase A from Serratia marcescens
J Biol Chem 295, 1915-1925 (2020) DOI:10.1074/jbc.RA119.012078 OA
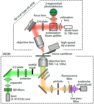
57. #Umakoshi T, #Fukuda S, Iino R, Uchihashi T, *Ando T (#Equal contribution)
High-speed near-field fluorescence microscopy combined with high-speed atomic force microscopy for biological studies
BBA General Subjects 1864, 129325 (2020) DOI:10.1016/j.bbagen.2019.03.011
2019
56. *Ando J, Nakamura A, Yamamoto M, Song C, Murata K, *Iino R
Multicolor high-speed tracking of single biomolecules with silver, gold, silver-gold alloy nanoparticles
ACS Photonics 6, 2870-2883 (2019) DOI:10.1021/acsphotonics.9b00953
Press release in EurekAlert
Optics & Photonics News: Nanoparticles Enable Multicolor Imaging of Biomolecules
Antpedia: 原来纳米粒子可以对生物分子进行多色成像
プレスリリース:光学顕微鏡によるマルチカラー高速高精度1分子観察を実現
オプトロニクスオンライン:分子研ら,光学顕微鏡でマルチカラー1分子観察 (2019年11月26日)
科学新聞:マルチカラー高速高精度1分子観察 分子研が光学顕微鏡で実現
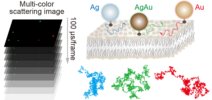
55. Iida T, Minagawa Y, Ueno H, Kawai F, Murata T, *Iino R
Single-molecule analysis reveals rotational substeps and chemo-mechanical coupling scheme of Enterococcus hirae V1-ATPase
J Biol Chem 294, 17017-17030 (2019) DOI:10.1074/jbc.RA119.008947 OA
Press release in EurekAlert
プレスリリース:回転分子モーターV1の化学力学エネルギー変換機構を解明
オプトロニクスオンライン:分子研ら,分子モーター機構をレーザーで解明(2019年10月28日)
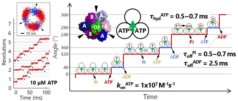
54. *Zhang Y, Minagawa Y, Kizoe H, Miyazaki K, Iino R, Ueno H, Tabata KV, Shimane Y, *Noji H
Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array
Sci Adv 5, eaav8185 (2019) DOI:10.1126/sciadv.aav8185 OA
Research introduction by Yi Zhang (JAMSTEC)
張さん(JAMSTEC)による研究紹介
2018
53. #Ando J, #Nakamura A, Visootsat A, Yamamoto M, Song C, Murata K, *Iino R (#Equal contribution)
Single-nanoparticle tracking with angstrom localization precision and microsecond time resolution
Biophys J 115, 2413-2427 (2018) DOI:10.1016/j.bpj.2018.11.016 OA
Press release in EurekAlert
プレスリリース:光学顕微鏡で原子レベルの位置決定精度を達成
科学新聞紹介記事(2018年12月14日1面、pdf 0.8MB)

52. Fujimoto K, Morita Y, Iino R, Tomishige M, Shintaku H, Kotera H, *Yokokawa R
Simultaneous observation of kinesin-driven microtubule motility and binding of adenosine triphosphate using linear zero-mode waveguides
ACS Nano 12, 11975–11985 (2018) DOI:10.1021/acsnano.8b03803
プレスリリース:モータータンパク質とATPの結合を1分子で可視化することに成功 ナノ構造に閉じ込めた光でモータータンパク質の仕組みに迫る
科学新聞紹介記事(2018年12月21日4面、pdf 1MB)

51. Tsunoda J, Song C, Lica Imai F, Takagi J, Ueno H, Murata T, Iino R, *Murata K
Off-axis rotor in Enterococcus hirae V-ATPase visualized by Zernike phase plate single-particle cryo-electron microscopy
Sci Rep 8, 15632 (2018) DOI:10.1038/s41598-018-33977-9 OA
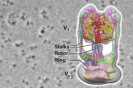
50. *Nakamura A, Okazaki K, Furuta T, Sakurai M, *Iino R
Processive chitinase is Brownian monorail operated by fast catalysis after peeling rail from crystalline chitin
Nat Commun. 9, 3814 (2018) DOI:10.1038/s41467-018-06362-3 OA
Press release in EurekAlert
プレスリリース:キチン加水分解酵素は熱ゆらぎを利用して一方向に動きながら結晶性バイオマスを分解する分子モノレールカーである
科学新聞紹介記事(2018年10月5日6面、pdf 1MB)
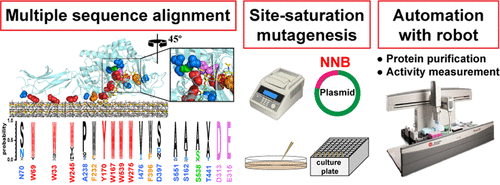
49. Kawai F, Nakamura A, Visootsat A, *Iino R
Plasmid-based one-pot saturation mutagenesis and robot-based automated screening for protein engineering
ACS Omega 3, 7715–7726 (2018) DOI: 10.1021/acsomega.8b00663 OA
48. #Uchihashi T, #*Watanabe YH, Nakazaki Y, Yamasaki T, Watanabe T, Maruno T, Ishii K, Uchiyama S, Song C, Murata K, *Iino R, *Ando T (#Equal contribution)
Dynamic structural states of ClpB involved in its disaggregation function
Nat Commun 9, 2147 (2018) DOI:10.1038/s41467-018-04587-w OA
Press release in EurekAlert
プレスリリース:凝集したタンパク質を再生する分子機械ClpB の動的な構造変化の可視化に成功
47. Nakamura A, Tasaki T, Okuni Y, Song C, Murata K, Kozai T, Hara M, Sugimoto H, Suzuki K, Watanabe T, Uchihashi T, Noji H, *Iino R
Rate constants, processivity, and productive binding ratio of chitinase A revealed by single-molecule analysis
Phys Chem Chem Phys 20, 3010-3018 (2018) DOI:10.1039/C7CP04606E
Featured on the inside back cover!
2016
46. #Baba M, #Iwamoto K, Iino R, Ueno H, Hara M, Nakanishia A, Kishikawa J, *Noji H, *Yokoyama K (#Equal contribution)
Rotation of artificial rotor axles in rotary molecular motors
Proc Natl Acd Sci USA 113, 11214-11219 (2016) DOI:10.1073/pnas.1605640113
45. Nakamura A, Tasaki T, Ishiwata D, Yamamoto M, Okuni Y, Visootsat A, Maximilien M, Noji H, Uchiyama T, Samejima M, Igarashi K, *Iino R
Single-molecule imaging analysis of binding, processive movement, and dissociation of cellobiohydrolase Trichoderma reesei Cel6A and its domains on crystalline cellulose
J Biol Chem 291, 22404-22413 (2016) DOI: 10.1074/jbc.M116.752048
Featured on the cover!
44. #Isojima H, #Iino R, Niitani Y, Noji H, *Tomishige M (#Equal contribution)
Direct observation of intermediate states during the stepping motion of kinesin-1
Nat Chem Biol 12, 290-297 (2016) DOI:10.1038/nchembio.2028
News and Views in Nature Chemical Biology: Motor proteins: Kinesin's gait captured
ブレスリリース「二本足で歩く分子モータータンパク質キネシンの足の動きを精細に可視化」
43. *Matsumoto Y, Sakakihara S, Grushnikov A, Kikuchi K, Noji H, Yamaguchi A, Iino R, Yagi Y, Nishino K
A microfluidic channel method for rapid drug-susceptibility testing of Pseudomonas aeruginosa
PLOS ONE 11, e0148797 (2016) DOI:10.1371/journal.pone.0148797
2015
42. Obayashi Y, Iino R, *Noji H
A single-molecule digital enzyme assay using alkaline phosphatase with a cumarin-based fluorogenic substrate
Analyst 140, 5065-73 (2015) DOI:10.1039/c5an00714c
41. Enoki S, Iino R, Niitani Y, Minagawa Y, Tomishige M, *Noji H
High-speed angle-resolved imaging of single gold nanorod with microsecond temporal resolution and one-degree angle precision
Anal Chem 87, 2079-2086 (2015) DOI:10.1021/ac502408c
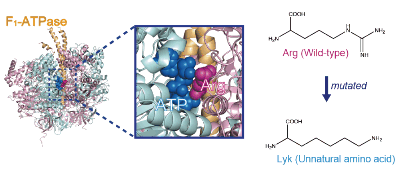
40. Yukawa A, Iino R, Watanabe R, Hayashi S, *Noji H
Key chemical factors of arginine finger catalysis of F1-ATPase clarified by an unnatural amino acid mutation
Biochemistry 54, 472–480 (2015) DOI:10.1021/bi501138b
2014
39. #Ueno H, #Minagawa Y, Hara M, Rahman S, Yamato I, Muneyuki E, Noji H, *Murata T, *Iino R (#Equal contribution)
Torque generation of Enterococcus hirae V-ATPase
J Biol Chem 289, 31212-31223 (2014) DOI:10.1074/jbc.M114.598177
38. Ikeda T, Tsukahara T, Iino R, Takeuchi M, *Noji H
Motion capture and manipulation of single synthetic molecular rotors by optical microscopy
Angew Chem Int Ed 53, 10082–10085 (2014) DOI:10.1002/anie.201403091
プレスリリース: 1ナノメートルの人工分子マシン1個を「見て、触る」ことに成功:光学顕微鏡による 1分子モーションキャプチャ
Will be featured on the back cover!
37. Ikeda T, Iino R, *Noji H
Real-time fluorescence visualization of slow tautomerization of single free-base phthalocyanines under ambient conditions
Chem Commun 50, 9443-9446 (2014) DOI:10.1039/C4CC02574A
マイナビニュース記事. Introduced in UTokyo Research! (English, Japanese)
Real-time visualization of slow tautomerization in a single organic chromophore : Candidate for a 1-nanometer-sized molecular memory
有機色素分子1個の遅い互変異性化をリアルタイムに記録:分子メモリ開発につながる可能性
Featured on the front cover!
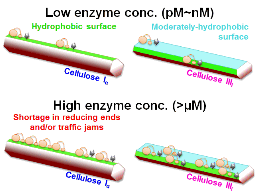
36. Shibafuji Y, Nakamura A, Uchihashi T, Sugimoto N, Fukuda S, Watanabe H, Samejima M, Ando T, Noji H, Koivula A, Igarashi K, *Iino R
Single-molecule imaging analysis of elementary reaction steps of Trichoderma reesei cellobiohydrolase I (Cel7A) hydrolyzing crystalline cellulose Iα and IIII
J Biol Chem 289, 14056-14065 (2014) DOI:10.1074/jbc.M113.546085
35. Takehara H, Miyazawa K, Noda T, Sasagawa K, Tokuda T, Kim SH, Iino R, Noji H, *Ohta J
A CMOS image sensor with stacked photodiodes for lensless observation system of digital enzyme-linked immunosorbent assay
Jpn J Appl Phys 53, 04EL02 (2014) DOI:10.7567/JJAP.53.04EL02
2013
34. #Minagawa Y, #Ueno H, Hara M, Ishizuka-Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Yamato I, Muneyuki E, Noji H, *Murata T, *Iino R (#Equal contribution)
Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase
J Biol Chem 288, 32700-32707 (2013) DOI:10.1074/jbc.M113.506329
Featured on the front cover! Designed by SCIEMENT
33. Fukuda S, Uchihashi T, Iino R, Okazaki Y, Yoshida M, Igarashi K, and *Ando T
High-speed atomic force microscope combined with single-molecule fluorescence microscope
Rev Sci Instrum 84, 073706 (2013) DOI:10.1063/1.4813280
32. Watanabe R, Tabata KV, Iino R, Ueno H, Iwamoto M, Oiki S, *Noji H
Biased Brownian stepping rotation of FoF1-ATP synthase driven by proton motive force
Nat Commun 4, 1631 (2013) DOI:10.1038/ncomms2631
2012
31. Enoki S, Iino R, Morone N, Kaihatsu K, Sakakihara S, Kato N, *Noji H
Label-free single-particle imaging of the influenza virus by objective-type total internal reflection dark-field microscopy
PLOS ONE 7, e49208 (2012) DOI:10.1371/journal.pone.0049208.
30. Kim SH, Iwai S, Araki S, Sakakihara S, Iino R, *Noji H
Large-scale femtoliter droplet array for digital counting of single biomolecules
Lab Chip 12, 4986-4991 (2012) DOI:10.1039/C2LC40632B
29. You H, Iino R, Watanabe R, *Noji H.
Winding single-molecule double-stranded DNA on a nanometer-sized reel
Nucleic Acids Research 40, e151 (2012) DOI:10.1093/nar/gks651
28. *Iino R, Hayama K, Amezawa H, Sakakihara S, Kim SH, Matsumoto Y, Nishino K, Yamaguchi A, Noji H.
A single-cell drug efflux assay in bacteria by using a directly accessible femtoliter droplet array
Lab Chip 12, 3923-3929 (2012) DOI:10.1039/C2LC40394C
Introduced as one of the Emerging Investigators in Lab on a Chip!
27. *Hayashi S, Ueno H, Shaikh AR, Umemura M, Kamiya M, Ito Y, Ikeguchi M, Komoriya Y, Iino R, Noji H.
Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single molecule observations
J Am Chem Soc 134, 8447–8454 (2012) DOI:10.1021/ja211027m
26. Komoriya Y, Ariga T, Iino R, Imamura H, Okuno D, *Noji H.
Principle role of the arginine finger in rotary catalysis of F1-ATPase
J Biol Chem 287, 15134-15142 (2012) DOI:10.1074/jbc.M111.328153
25. *Sasagawa K, Ando K, Kobayashi T, Noda T, Tokuda T, Kim SH, Iino R, Noji H, Ohta J.
Complementary metal–oxide–semiconductor image sensor with microchamber array for fluorescent bead counting
Jpn J Appl Phys 51, 02BL0 (2012) DOI:10.1143/JJAP.51.02BL01
24. #Watanabe R, #Okuno D, Sakakihara S, Shimabukuro K, Iino R, Yoshida M, *Noji H. (#Equal contribution)
Mechanical modulation of catalytic power on F1-ATPase
Nat Chem Biol 8, 86-92 (2012) DOI:10.1038/nchembio.715
2011
23. #Uchihashi T, #Iino R, *Ando T, *Noji H. (#Equal contribution)
High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase
Science 333, 755-758 (2011) DOI:10.1126/science.1205510
Perspective of the Science
News of the Chemistry World
Research Highlight of the Nature Nanotech.
News of the BioTechniques
22. *Matsumoto Y, Hayama K, Sakakihara S, Nishino K, Noji H, *Iino R, Yamaguchi A.
Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels
PLOS ONE 6, e18547 (2011)
2010
21. Okuno D, Iino R, and *Noji H.
Stiffness of γ subunit of F1-ATPase
Eur Biophys J 39:1589-1596 (2010)
20. Watanabe R, Iino R, *Noji H.
Phosphate-release in F1-ATPase catalytic cycle follows ADP release
Nat Chem Biol 6, 814–820 (2010) DOI:10.1038/nchembio.443
Introduced in News and Views!
19. *Hayashi K, Ueno H, Iino R, and *Noji H.
Fluctuation theorem applied to F1-ATPase
Phys Rev Lett 104, 218103-1-218103-4 (2010)
18. Ueno H, Nishikawa S, Iino R, Tabata KV, Sakakihara S, Yanagida T, *Noji H.
Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution
Biophys J 98, 2014-2023 (2010)
17. Saita E, Iino R, Suzuki T, Feniouk BA, Kinosita K. Jr, *Yoshida M.
Activation and stiffness of the inhibited states of F1-ATPase probed by single-molecule manipulation
J Biol Chem 285, 11411-11417 (2010)
16. Sakakihara S, Araki S, *Iino R, *Noji H.
A single-molecule enzymatic assay in a directly accessible femtoliter droplet array
Lab Chip 10, 3355-3362 (2010)
2009
15. *Imamura H, Huynh Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai,T, *Noji H.
Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators
Proc Natl Acad Sci USA 106, 15651-15656 (2009)
14. Enoki S, Watanabe R, Iino R, and *Noji H.
Single-molecule study on the temperature-sensitive reaction of F1-ATPase with a hybrid F1 carrying a single β(E190D)
J Biol Chem 284, 23169-23176 (2009)
13. *Iino R, Hasegawa R, Tabata KV, *Noji H.
Mechanism of inhibition by C-terminal α-helices of the ε subunit of Escherichia coli FoF1-ATP synthase
J Biol Chem 284, 17457-17464 (2009)
2008
12. Okuno D, Fujisawa R, Iino R, Hirono-Hara Y, Imamura H, *Noji H.
Correlation between the conformational states of F1-ATPase as determined from its crystal structure and single-molecule rotation
Proc Natl Acad Sci USA 105, 20722-20727 (2008)
11. Watanabe R, Iino R, Shimabukuro K, Yoshida M, *Noji H.
Temperature-sensitive reaction intermediate of F1-ATPase
EMBO rep 9, 84-90 (2008)
2007
10. Suzuki KGN, Fujiwara TK, Sanematsu F, Iino R, Edidin M, and *Kusumi A.
GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1
J Cell Biol 177, 717-730 (2007)
2005
9. Iino R, Murakami T, Iizuka S, Kato-Yamada Y, Suzuki T and *Yoshida M.
Real-time monitoring of conformational dynamics of the epsilon subunit in F1-ATPase
J Biol Chem 280, 40130-40134 (2005)
8. Makiyo H, Iino R, Ikeda C, Imamura H, Tamakoshi M, Iwata M, Stock D, Bernal RA, Carpenter EP, Yoshida M, *Yokoyama K and *Iwata S.
Structure of a central stalk subunit F of Prokaryotic V-type ATPase/synthase from Thermus thermophilus
EMBO J 24, 3974-3983 (2005)
7. Koyama-Honda I, Ritchie K, Fujiwara T, Iino R, Murakoshi H, Kasai RS, and *Kusumi A.
Fluorescence imaging for monitoring the colocalization of two single molecules in living cells
Biophys J 88, 2126-2136 (2005)
2004
6. Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino R, Yamashita H, Saito M, Murakoshi H, Ritchie K, and *Kusumi A.
Ultrafine membrane Compartments for molecular diffusion as revealed by single molecule techniques
Biophys J 86, 4075-4093 (2004)
5. Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, and *Kusumi A.
Single-molecule imaging analysis of ras activation in living cells
Proc Natl Acad Sci USA 101, 7317-7322 (2004)
2003
4. Suzuki T, Murakami T, Iino R, Suzuki J, Ono S, Shirakihara Y, and *Yoshida M.
FoF1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of epsilon subunit in response to proton motive force and ADP/ATP balance
J Biol Chem 278, 46840-46846 (2003)
3. Nakada C., Ritchie K., Oba Y, Nakamura M, Hotta Y, Iino R, Kasai S, Yamaguchi K, Fujiwara T, and *Kusumi A.
Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization
Nat Cell Biol 5, 626-632 (2003)
2. Ike H, Kosugi A, Kato A, Iino R, Hirano H, Fujiwara T, Ritchie K, and *Kusumi A.
Mechanism of Lck recruitment to the T-cell receptor cluster as studied by single molecule fluorescence video imaging
ChemPhysChem 4, 620-626 (2003).
2001
1. Iino R, Koyama I, and *Kusumi A.
Single molecule imaging of green fluorescent protein in living cells: E-cadherin forms oligomers on the free cell surface
Biophys J 80, 2667-2677 (2001)