The 26th CIMoS Seminar
Molecular simulations of RNA folding
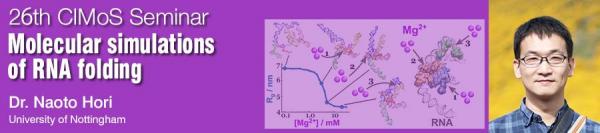
RNA molecules play a variety of functional roles in the cell. In considering such molecular mechanism, it is necessary to understand how the tertiary structure of RNA is formed and dynamically changed. For example, ribozymes need to be folded into specific compact structures for their catalytic activity; some messenger RNA and long non-coding RNAs also use specific structures for regulating translation and molecular recognition. Using coarse-grained molecular simulations, we have studied RNA folding under various solution conditions. Although RNA folding is mainly driven by the formation of base pairs and base stacks, it is also largely affected by ions, particularly divalent cations such as Mg, because RNA molecules are highly negatively charged. In this seminar, I will introduce our molecular simulation approach and show some results of RNA folding and misfolding, focusing on the interaction with ions.
References
Roca, Hori, et al., Proc. Natl. Acad. Sci. U.S.A. (2018) 10.1073/pnas.1717582115
Hori, Denesyuk, Thirumalai, Biophys. J. (2019) 10.1016/j.bpj.2019.04.037
Nguyen, Hori, Thirumalai, Proc. Natl. Acad. Sci. U.S.A. (2019) 10.1073/pnas.1911632116
Hori, Denesyuk, Thirumalai, Proc. Natl. Acad. Sci. U.S.A. (2021) 10.1073/pnas.2020837118
Nguyen, Hori, Thirumalai, Nat. Chem. (2022) 10.1038/s41557-022-00934-z
| Date | Thursday, 8th September, 2022 16:30 - 17:30 |
|---|---|
| Place | Room 201, IMS main office building (onsite) & Zoom meeting (online) |
| Title | Molecular simulations of RNA folding |
| Speaker | Naoto Hori School of Pharmacy, University of Nottingham |

